Bacterial inclusion bodies have functional proteins
At the end of 70ths, the recombinant DNA technology was developed and, with this, it began the production of protein of interest in easily handily organisms and its commercialization by a wide number of pharmacological companies. These industries use mainly the bacteria as producer organisms, since these allow to design fast and low cost production processes. However, even the evident advantages that presents the use of these organisms, nowadays there is still an important obstacle that it is necessary to overcome: protein aggregation.
The aggregation is a phenomenon that appears when bacteria are under cellular stress, such us the one that occurs when we force them to produce the protein of interest at high concentrations. As a result of the stress situation, the insoluble protein produced accumulates as aggregates called inclusion bodies. Inclusion bodies have been described like highly hydrated, resistant dense protein aggregates, refractile, cylindrical and with variable size. In general, it has been believed that the protein embedded in these aggregates is inactive and this fact has seriously restricted the spectrum of proteins marketed by the biotechnology industry, since the inactive form is useless.
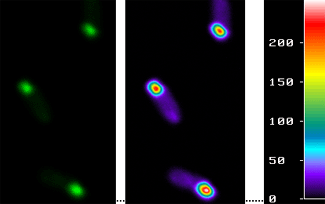
In our two last works (Garcia-Fruitos et al.;Garcia-Fruitos, Aris, and Villaverde), we have demonstrated that the protein present in many of these aggregates found in bacteria is not only active, but that are in the core of these aggregates, following a defined pattern. It was already known that inclusion bodies contains protein with an amyloid-like structure and this structure is believed to be the main cause of some human disorders such as Alzheimer's disease.
However, we have described the coexistence of both active and inactive proteins, a fact that leads to a total change in the general model that described until now inclusion bodies biology. To perform these experiments we have used a green fluorescent protein, since it is easily visualizable by fluorescence microscopy. In fact, this finding can mean an important change in the biotechnological field, since inclusion bodies can be used as biocatalysts, when they are formed by enzymes.
References
-Garcia-Fruitos, E., A. Aris, and A. Villaverde. "Localization of functional polypeptides in bacterial inclusion bodies". Appl.Environ.Microbiol. 73.1 (2007): 289-94.
- Garcia-Fruitos, E., et al. "Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins". Microb.Cell Fact. 4 (2005): 27.

